Environmental Engineering and Environmental Science
World-renowned faculty, state-of-the-art facilities, and innovative teaching and research prepare students to positively impact the world by addressing problems in the built and natural environments—discover environmental and water resources engineering and change the world around you.
The Environmental Engineering and Science laboratories include over 10,000 square feet of space devoted to environmental research activities. Instrumentation is currently available to perform analytical measurements associated with research investigations in water quality, air quality, solid and hazardous wastes, and environmental microbiology. Laboratory facilities are rated for Biosafety Level 1 and 2 work.
Major equipment includes: infrared, visible, and ultraviolet spectrophotometers; chemical fume hoods; a suite of environmental field sensors; a HIAC/Royco Model 9703 liquid particle counter; a Perkin Elmer atomic absorption spectrophotometer (AAnalyst 300 with autosampler/autodilution); a Thermo Scientific refrigerated Legend XFR centrifuge; a Beckman Coulter XPN 100 refrigerated ultracentrifuge; an Eppendorf Vacufuge; three 2 L B.Braun Biostat i fermentors, a 3 L Applikon fermentor; a 3 L New Brunswick Bioflo 3000 fermentor; apparatus for gel electrophoresis; a 400 Watt sonicator for cell fractionation; carbon, hydrogen, nitrogen, and oxygen analyzers for solid materials; four shaker table facilities; Zeiss UEM research microscope with phase contrast, DIC, and epifluorescence capabilities; a Dohrmann DC-80 TOC Analyzer; six gas chromatographs with ECD (2), FID (2), AFID, and TC detectors; two gas chromatographs with mass selective detectors (GC/MSD) (Varian Saturn 2000 and Agilent Technologies 5973); Dionex ICS 2000 ion chromatographs with autosampler; Coy anaerobic chambers; Milestone DMA direct mercury analyzer, four temperature controlled rooms; a PCR thermocycler and associated equipment including a hybridization oven; and a Kodak image analysis systems.
Environmental Laboratories
The City of Lawrence wastewater treatment plant has a secondary treatment capacity of 25 million gallons per day (MGD), and an average annual flow rate of 9 MGD. The plant performs nitrification to meet a daily ammonia limit of 7.5 mg/L, but there is no current discharge limit for total nitrogen and phosphorus. The City has historically hosted pilot wastewater systems for Dr. Sturm’s research, providing access to wastewater and power to operate the system. University employees follow all City Utility and OSHA safety requirements while on City property.

The Environmental Research and Environmental Instrument Laboratories are located in Learned Hall and LEEP2. The Learned Hall labs were renovated in 2007. The main lab comprises 2600 sq. ft. of wet lab, analytical chemistry, microbiology, and fluorescent microscopy workspaces. Dr. Hutchison maintains a 572 sq. ft. Biosafety Level 2 lab. The LEEP2 lab includes 2433 sq. ft. of wet lab space. Dr. Sturm also oversees two labs (a 450 sq. ft Greenhouse and a 730 sq. ft. wet lab) in the Measurement, Materials and Sustainable Environment Center, which was dedicated in fall 2012. In total, these laboratories contain all the equipment necessary to perform reactor and biological growth experiments. The School of Engineering has a full-time technician staff and machine shop to help design, build, and maintain experimental setups. A list of key research capabilities for this space is provided below:
- Biological Reactor Studies (bench-scale)
- Sequencing Batch Reactors for Wastewater Treatment
- Granular Activated Sludge reactors
- Biofilm reactors and biofilm characterization
- Algal Photobioreactors
- Anaerobic reactors
- Treatability Studies (microcosms, bench-scale, mesocosms)
- Testing of biological treatment capacity at multiple scales
- Chemical Analysis
- UV-Spectrophotometry methods
- Gas Chromatography – Mass Spectrometry
- High Performance Liquid Chromatography
- Ion Chromatography
- Total Organic Carbon and Nitrogen
- Metals and Elemental Analysis (CHONS, AA)
- Biological Analysis
- DNA extraction and quantification
- quantitative PCR
- Fluorescent microscopy for species identification (FISH)
- Extracellular polymeric substance studies (chemical and microscopic)
- Metagenomic analysis using Illumina sequencing
- Protein/enzyme extraction
- Enzymatic analysis
The University of Kansas Field Station Nelson Environmental Study Area (NESA) is located within the larger University of Kansas Field Station & Ecological Reserves (KSR), which totals more than 3,000 acres of habitats, experimental tracts, and support facilities. The University of Kansas Field Station has served a prominent role in ecological, environmental, and toxicological research for more than half a century.
NESA is devoted to experimental ecological studies, including the Kansas Aquatic Mesocosm Program, which includes more than 100 experimental ponds and aquatic enclosures, as well as Frank B. Cross Reservoir, a protected watershed. As the headquarters for the larger ecological reserves, NESA also houses a year-round caretaker residence, a meteorological station, an Aquatic Laboratory, the KSR Headquarters, overnight cabins, a well-equipped shop, maintenance and storage buildings, vehicles, and heavy equipment. The original headquarters building was constructed in 1994 with the aid of an NSF grant. In the last five years, another NSF grant has allowed for expansion of the headquarters, with a 2700 sq. ft. addition, including the 1700 sq. ft. Armitage Education Center. This includes a large classroom/meeting room, two laboratories, a lobby/great room, a full kitchen, offices, and shower and laundry facilities.
In addition to these excellent facilities, three full-time technical and maintenance staff are available at NESA to maintain the greater field station and to aid experimental operations.
The Center for Metagenomic Microbial Community Analysis is a multidisciplinary initiative comprising faculty from multiple departments and fields conducting research from genes to ecosystems. The Center was established as an internal investment by the University of Kansas and supports a full-time PhD-level Research Associate and a Master's-level Research Assistant. Staff perform metagenomic analysis of microbial communities from a range of environmental samples. With the support of the Genome Sequencing Core and the Advanced Computing Facility, high-throughput sequencing through computationally intensive data analysis is performed.
The Genome Sequencing Core lab is part of the Center for Molecular Analysis of Disease Pathways, an NIH-funded Center for Biomedical Research Excellence (COBRE) infrastructure grant. The Core’s mission is to provide state-of-the-art next generation sequencing capabilities and to serve as a catalyst for interaction among genomics research facilities, building a strong foundation for genomics research at KU. The Core has two full-time staff members in addition to the core leader, Dr. Erik Lundquist. The facility provides an Illumina HiSeq 2500 platform and charges user fees at a competitive price.
The Advanced Computing Facility was expanded in 2013 with a $4.7 million grant from the National Center for Research Resources at NIH, and a major infrastructure improvement grant from NSF. Dan Voss, the newly hired Director of Research Computing, works with researchers to develop interfaces between the Genome Sequencing Core, where sequencing data is generated, and the Community Cluster, where it is processed. The Center for Microbial Metagenomics Community Analysis has purchased four nodes in the Cluster for general bioinformatics data analysis.
 Algal Biofuel Production
Algal Biofuel Production
Mesocosms were operated at the Nelson Environmental Studies Area to operate as a continuous flow system, fed with municipal wastewater to cultivate algal biomass for biofuel studies.
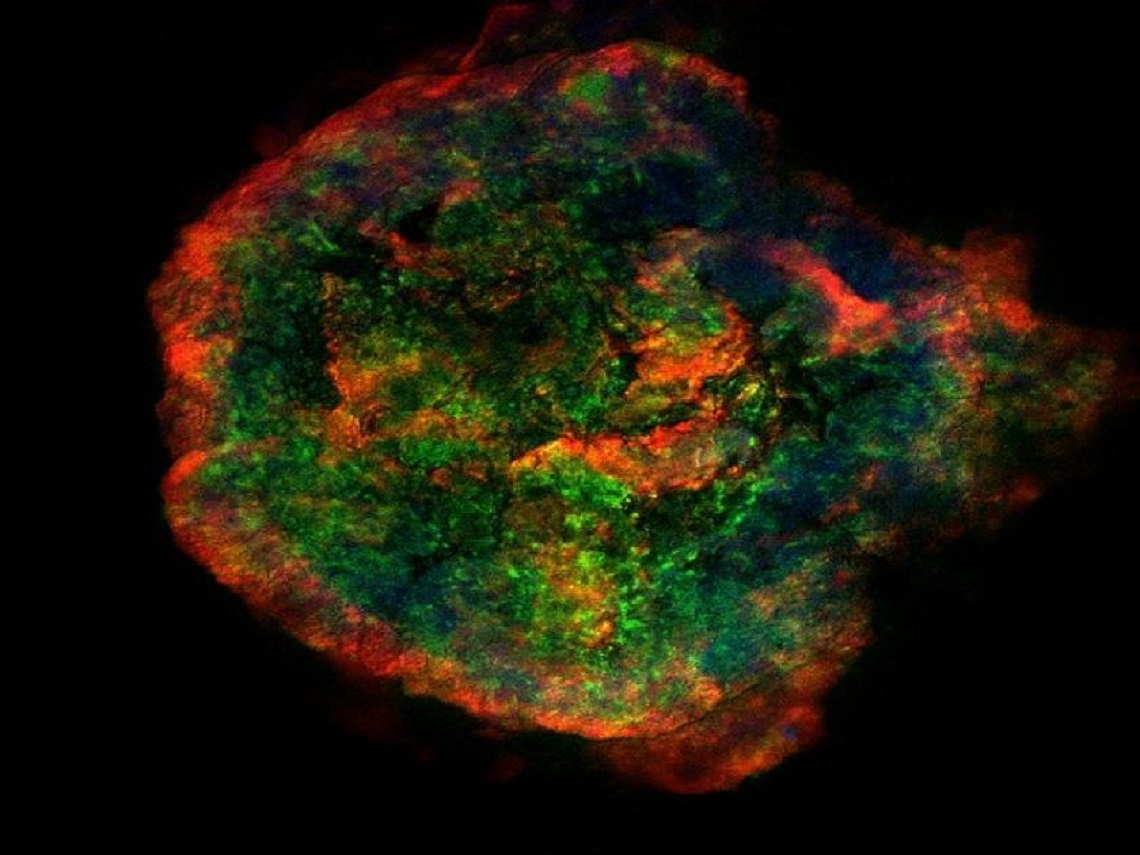
Studying Biofilm Structure
An aerobic granule is fluorescently stained with protein, carbohydrate, and DNA stains to visualize the cellular and extracellular polymeric substances structure of the biofilm.

Climate Change and Microbial Cycling
Linking soil carbon and nitrogen transformations with climate change.

Restoring Fungi in Degraded Sites
A mushroom breaks through the sandy soils of a disturbed scrub site in central FL. We are analyzing soil fungi, then using additions of native fungi to try to accelerate restoration.

Biological Wastewater Treatment
Bench-scale sequencing batch reactors were used to treat municipal wastewater at increasing sludge residence times. The experiment tested how increased SRT affects community diversity. Increasing diversity has been shown to increase the removal of some pharmaceuticals, such as estrogen (EE2).

Fate of Antibiotic Resistance Genes
Mesocosm experiments were performed at the Nelson Environmental Studies Area (NESA) to test the fate of antibiotic resistance genes from swine manure. Mesocosms were covered with varying degrees of shade cloth to control light penetration into the mesocosms.
The Microscopy & Analytical Imaging Laboratory at the University of Kansas is a common use service lab. The laboratory recently underwent extensive renovations and has acquired a wide spectrum of analytical imaging devices with very flexible imaging capabilities. Within the lab, a wide range of optics, high speed shutters, and spectral analysis tools are also available, permitting flexible configurations of devices for specialized applications including, but not limited to, total internal reflection fluorescence microscopy, spectral confocal microscopy (laser scanning confocal microscopy platform), spinning disk confocal microscopy, measurement of multi-spectral polarization anisotropy, dynamic measurements of fluorescent dipole orientation, fluorescence correlation spectroscopy, and high speed FRET detection. The laboratory is also home to a 120keV TEM, a Field Emission SEM equipped with EDS and Cathodoluminescence detection, a fluorescence 2-D gel/microtitre plate/microarray scanning system and an AFM configured for both biological and materials analysis.
Of particular note for biological samples, a Zeiss Meta 510, multi-line laser scanning confocal microscope is available in either an upright configuration or on an inverted microscope platform; the inverted platform possesses multi-well plate imaging capabilities, advanced spectroscopic analysis functions, laser micro-dissection, and advanced FRET/FLIP/FRAP capabilities. All of the functions of this microscope may be automated through an open, visual basic scripting environment. An additional pair of optical microscopes, one inverted, one upright, are available for standard, non-laser based epi-fluorescence imaging. All optical microscopes in the lab are equipped with phase contrast and DIC optics, automated objective/filter set switching, computer driven Z -axis motor drives, and auto-focusing under software control.